Abstract
PURPOSE Worldwide clinical knowledge is expanding rapidly, but physicians have sparse time to review scientific literature. Large language models (eg, Chat Generative Pretrained Transformer [ChatGPT]), might help summarize and prioritize research articles to review. However, large language models sometimes “hallucinate” incorrect information.
METHODS We evaluated ChatGPT’s ability to summarize 140 peer-reviewed abstracts from 14 journals. Physicians rated the quality, accuracy, and bias of the ChatGPT summaries. We also compared human ratings of relevance to various areas of medicine to ChatGPT relevance ratings.
RESULTS ChatGPT produced summaries that were 70% shorter (mean abstract length of 2,438 characters decreased to 739 characters). Summaries were nevertheless rated as high quality (median score 90, interquartile range [IQR] 87.0-92.5; scale 0-100), high accuracy (median 92.5, IQR 89.0-95.0), and low bias (median 0, IQR 0-7.5). Serious inaccuracies and hallucinations were uncommon. Classification of the relevance of entire journals to various fields of medicine closely mirrored physician classifications (nonlinear standard error of the regression [SER] 8.6 on a scale of 0-100). However, relevance classification for individual articles was much more modest (SER 22.3).
CONCLUSIONS Summaries generated by ChatGPT were 70% shorter than mean abstract length and were characterized by high quality, high accuracy, and low bias. Conversely, ChatGPT had modest ability to classify the relevance of articles to medical specialties. We suggest that ChatGPT can help family physicians accelerate review of the scientific literature and have developed software (pyJournalWatch) to support this application. Life-critical medical decisions should remain based on full, critical, and thoughtful evaluation of the full text of research articles in context with clinical guidelines.
- artificial intelligence
- large language models
- ChatGPT
- primary care research
- critical assessment of scientific literature
- bias
- text mining
- text analysis
INTRODUCTION
Nearly 1 million new journal articles were indexed by PubMed in 2020, and worldwide medical knowledge now doubles approximately every 73 days.1 Meanwhile, care models emphasizing clinical productivity2,3 leave clinicians with scant time to review the academic literature, even within their own specialty.
Recent developments in artificial intelligence (AI) and natural language processing might offer new tools to confront this problem. Large language models (LLMs) are neural network–based computer programs that use a detailed statistical understanding of written language to perform many tasks including text generation, summarization, software development, and prediction.4-12 One LLM, Chat Generative Pretrained Transformer (ChatGPT) has recently garnered substantial attention in the popular press.13-17 We wondered if LLMs could help physicians review the medical literature more systematically and efficiently.
Unfortunately, LLMs can also “hallucinate,” producing text that, whereas often convincing and seemingly authoritative, is not fact based.18-21 In addition, many concerns have been raised regarding the possibility of bias in AI models including LLMs. Bias in AI models can arise from both implicit and explicit biases present in their training data sets.22,23 Additional biases might potentially arise during the fine-tuning process. Large language models can be fine-tuned via a reinforcement learning approach, which uses feedback from humans to improve model performance.9 Such feedback might carry implicit and/or explicit biases of the humans providing feedback. Responsible use of LLMs at any stage of the clinical research process therefore requires careful validation to ensure that specific uses are unlikely to exacerbate preexisting systemic inequalities in health care.
To perform tasks, LLMs are prompted with instructions and supporting information. We wondered if LLMs—when carefully instructed—could (1) help clinicians find articles relevant to their medical specialty and (2) produce reasonable summaries of the major findings without introducing inaccuracies as a result of hallucination. Specifically, we investigated whether ChatGPT-3.5 could produce (1) high quality, (2) accurate, and (3) bias-free summaries of medical abstracts, focusing on points that were most likely to be salient for practicing physicians. We also prompted ChatGPT to self-reflect on the quality, accuracy, and biasness of its own summaries and evaluated its performance in classifying articles’ relevance to various medical specialties (eg, internal medicine, surgery, etc). Self-reflections have been used to improve the ability of LLMs to perform logical reasoning.24 We compared these self-reflections and relevance classifications to annotations by human physicians.
METHODS
Article Selection
We analyzed 10 articles from each of 14 selected journals (Table 1). These journals were chosen to (1) include topics ranging across medicine, (2) include both structured and unstructured abstracts in the sample, and (3) span a large range of journal impact factors. We drew articles from research articles published in 2022 by simple random sampling of each journal. ChatGPT was trained on data assembled before 2022; therefore, we reasoned that these articles would not have been included in the training corpus (ie, not seen by ChatGPT previously). We included case series, observational studies, interventional studies, randomized controlled trials, systematic reviews, and meta-analyses. We excluded editorials, letters, perspectives, errata, nonsystematic reviews, and single-case reports.
Attributes of Journals Selected for Analysis
ChatGPT Prompt and Data Extraction
We prompted ChatGPT with instructions (Supplemental Appendix 1) to summarize the abstract, self-reflect on the quality, accuracy, and degree of bias present in its summary, and classify the relevance of the abstract to 10 areas of medicine (cardiology, pulmonary medicine, family medicine, internal medicine, public health, primary care, neurology, psychiatry, obstetrics and gynecology, and general surgery). We instructed ChatGPT to adhere to a word limit of 125 words, but no efforts were otherwise undertaken to enforce this directive. Quality, accuracy, bias, and relevance were all evaluated on scales of 0-100. ChatGPT was given the full text of the abstract (as of February 2022 in PubMed) but was not provided with any other metadata (eg, journal, publication date) for the articles. We transcribed the Chat-GPT-produced summary, along with the quality, accuracy, bias, and relevance scores, into Research Electronic Data Capture (REDCap; project-redcap.org) for data management.25,26 We used the ChatGPT-3.5 (standard) model as of February 2022.
Physician Evaluation of ChatGPT Summaries
Seven physicians independently reviewed summaries. For each article, physician reviewers received the article’s title, journal, PubMed ID, abstract, and the GPT-produced summary of the article via REDCap. The reviewers classified the quality, accuracy, and amount of bias present in the summaries on a 0-100 scale. Reviewers also evaluated the relevance of the articles to various areas of medicine on a 0-100 scale. To harmonize scores, reviewers used a common rubric to assign scores. For quality and accuracy, reviewers scored these on a 0-100 scale with anchors on the typical letter grade (A, B, C, D, or F) in the common American grading system, with corresponding ranges of 90-100, 80-89, 70-79, 60-69, and ≤59. For bias, reviewers used a common rubric ranging from “no bias” to “blatantly biased” (Supplemental Appendix 1). Reviewers also determined if the summary contained any evidence of bias on the basis of race, color, religion, sex, gender, sexual orientation, or national origin that was not present in the abstract. For relevance, reviewers were instructed to use a common rubric ranging from “clearly relevant” to “not relevant” (Supplemental Appendix 1). Each reviewer evaluated approximately 45 summaries. Reviews were randomly distributed to reviewers, and every summary was reviewed by 2 reviewers. The first 5 completed reviews for each reviewer were considered part of the reviewer burn-in phase and discarded. The review team comprised individuals of varied sexes, genders, races, religions, and national origins. The senior author (D.P.) acted as referee for the other reviewers and therefore did not review abstracts and summaries. Scores for quality, accuracy, bias, and relevance were averaged across all reviewers to produce final quality, accuracy, bias, and relevance scores for each summary. Reviewers also annotated both minor and serious factual inaccuracies. Serious factual inaccuracies were those that would change a major interpretation of an article. When substantial factual inaccuracies or biases were noted, reviewers supplied a text description of them.
Statistical and Qualitative Analyses
We performed statistical analyses using R version 4.2.2 (The R Foundation; r-project.org). To evaluate quality, accuracy, and bias, we calculated descriptive statistics (1) for the overall sample and (2) stratified by journal. We qualitatively compared the quality, accuracy, and bias score distributions for (1) ChatGPT, (2) individual human reviewers, and (3) all human reviewers in aggregate using violin plots and scatterplots.
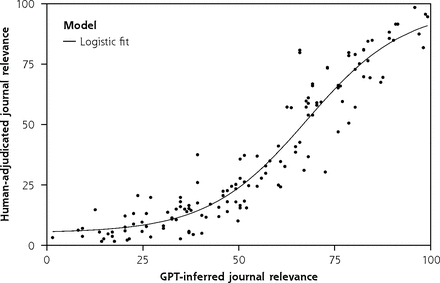
For scores assigned by ChatGPT and human reviewers on how related each article was to various medical specialties, we conducted analyses at 2 levels: journal level and article level. First, we analyzed agreement at the journal level between the ChatGPT-assigned relevance scores with (1) a priori expectations, and (2) human scores. For this journal-level analysis, we averaged across all articles for a given journal. For example, the “relevance to public health” score for Annals of Family Medicine is the average of all “relevance to public health” scores for all Annals of Family Medicine articles included. We expected a monotonic—although not necessarily linear—relation between the ChatGPT and human relevance scores. We could have asked ChatGPT and humans to make a dichotomous relevant/not relevant determination for each article. Instead, we collected more granular data on a scale of 0 (not relevant) to 100 (very relevant). In analogy to logistic regression analysis of categorical variables for classification, we modeled the nonlinear relation between ChatGPT (x) and human (y) relevance scores at the journal level using a 4-variable logistic function: y ~C + L/[1+e−k(x-x)], where L models the difference in maximal and minimal human scores, x0 models the difference in leniency between ChatGPT and humans, C is related to the difference in mean human score and mean ChatGPT score, and k describes the linear slope of the relation between human and ChatGPT scores near the midpoint of the fit. Nonlinear fits were computed using the nls function in the base R Stats package.
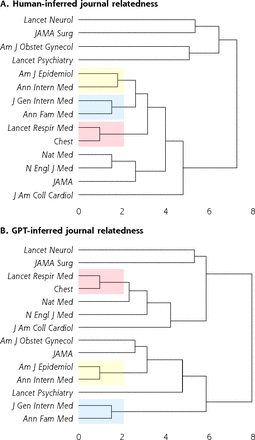
In addition, we defined the relevance profile for each journal as the vector of relevance scores for each specialty assigned to that journal. By this method, each journal received a ChatGPT-estimated relevance profile and a human-estimated relevance profile. The Euclidean distance between the relevance profile of 2 journals estimates their content similarity. We hierarchically clustered the journals via an agglomerative approach using both the ChatGPT and human relevance profiles and qualitatively compared the clustering dendrograms implied by ChatGPT- and human-assigned relevance scores.
At the article level, we evaluated the relation between ChatGPT relevance scores and human relevance scores (1) across all specialties in aggregate and (2) stratified by specialty. For the aggregate analysis, we again performed nonlinear regression with the 4-variable logistic function. A sensitivity analysis using a nonlinear mixed model, including reviewer identities as a random effect, was explored but did not substantially improve the quality of the nonlinear fit. For the analyses stratified by specialty, we used linear regression and calculated the coefficient of determination (R2) for ChatGPT-predicted vs human-assigned scores within each specialty.
We used the Kruskal-Wallis rank-sum test to evaluate for differences in quality, accuracy, and bias scores stratified by (1) journal of origin and (2) structured vs unstructured abstracts.
Human Subjects Protection
This project was determined to be Not Human Subjects Research by the University of Kansas Medical Center Institutional Review Board.
RESULTS
Characteristics of ChatGPT Summaries of Medical Abstracts
We used ChatGPT to summarize 140 abstracts across 14 journals (Table 1). Most abstracts (n = 120) used a structured format. Abstracts included a mean of 2,438 characters. The ChatGPT summaries decreased this length by 70% to a mean of 739 characters. An example summary produced by Chat-GPT is shown in Supplemental Appendix 2.
Summaries were scored by physician reviewers as high quality (median score 90.0, interquartile range [IQR] 87.0-92.5), high accuracy (median 92.5, IQR 89.0-95.0), and low bias (median 0, IQR 0-7.5) (Table 2). ChatGPT’s self-reflections also rated the summaries as high quality, high accuracy, and low bias, concordant with the judgements of human evaluators. We found no difference in human-assigned scores when stratifying by (1) journal of origin or (2) structured vs unstructured abstracts.
Median Quality, Accuracy, and Bias Scores Assigned by Humans and ChatGPT to Articles Overall and Stratified by Journal
Hallucinations and Inaccuracies
We qualitatively annotated instances of serious inaccuracies and hallucinations—defined as those that changed major interpretation of a study—in 4 of 140 summaries. One of these cases was due to omission; a significant risk factor (female gender) was found using a logistic regression model and was omitted from the summary whereas all other significant risk factors were reported. One of these cases was the result of apparent misunderstanding by ChatGPT of the semantic meaning of the abstract; a complicated study included 2 treatment arms with different primary outcomes, but the summary implied that the arms had the same primary outcomes. Two cases were due to hallucination; 1 summary stated that a randomized trial was double-blinded when the abstract clearly stated that it was open label, and 1 summary stated that results were consistent across subgroups, but only 1 of the many endpoints evaluated in that study was reported for the subgroup in the abstract. Minor inaccuracies were noted in 20 of 140 articles, related to the introduction of ambiguity in meaning (n = 2) or summarization away of details that would have provided additional content but not completely change the meaning (n = 18), for example, in cases in which an effect size was statistically significant but of questionable clinical significance.
ChatGPT Annotations of Journal Relevance Compared to Human Annotations
We included journals spanning many medical specialties. Our hypothesis was that ChatGPT would be able to classify articles drawn from a given journal as relevant to that journal’s topical focus. For example, we would expect The Lancet Neurology to have high relevance to neurology and low relevance to obstetrics and gynecology. At the aggregate level, this hypothesis was borne out; the ChatGPT relevance profile of journals agreed with a priori expectations (Table 3). Likewise, there was a strong nonlinear association between physician and ChatGPT relevance scores at the journal level (standard error of the regression 8.6) (Figure 1). The standard error of the regression is related to the typical expected prediction error for the regression model. On our relevance scale, ranging from 0 to 100, a standard error of the regression of 8.6 can therefore be interpreted as an 8.6% expected error in the predicted human-assigned relevance given the ChatGPT-assigned relevance. Clustering analysis (Figure 2) revealed similarity of the general structure of the clustering dendrograms in both the ChatGPT and human dendrograms. General medicine and cardiovascular medicine journals cluster together, with specialist journals (psychiatry, neurology) branching out from the root of the tree. Likewise, 3 of the 4 strongest associations (American Journal of Epidemiology with Annals of Internal Medicine, Journal of General Internal Medicine with Annals of Family Medicine, The Lancet Respiratory Medicine with Chest) inferred from the human relevance profiles were also present in the ChatGPT dendrogram (Figure 2; red-, yellow-, and blue-shaded areas). We thus conclude that ChatGPT can infer the relevance profile of journals based on analysis of the abstracts of its articles.
Journal Relevance Profiles; Topical Content of Each Journal as Annotated by ChatGPT
Agreement between human and GPT relevance scores at the journal level.
GPT = Generative Pretrained Transformer.
Clustering dendrograms for journal relatedness.
Am J Epidemiol = American Journal of Epidemiology; Am J Obstet Gynecol = American Journal of Obstetrics and Gynecology; Ann Fam Med = Annals of Family Medicine; Ann Intern Med = Annals of Internal Medicine; ChatGPT = Chat Generative Pretrained Transformer; GPT = Generative Pretrained Transformer; J Am Coll Cardiol = Journal of the American College of Cardiology; JAMA = Journal of the American Medical Association; JAMA Surg = JAMA Surgery; J Gen Intern Med = Journal of General Internal Medicine; Lancet Neurol = The Lancet Neurology; Lancet Psychiatry = The Lancet Psychiatry; Lancet Respir Med = The Lancet Respiratory Medicine; N Engl J Med = The New England Journal of Medicine; Nat Med = Nature Medicine.
Note: Human (panel A) and ChatGPT (panel B) dendrograms are shown. Strong relations between journals that are preserved in both dendrograms are highlighted in blue, yellow, and red.
Ability of ChatGPT to Classify Relevance of Individual Articles to Various Disciplines of Medicine
We next evaluated whether ChatGPT could classify the relevance of individual articles. Within individual specialties, the relation between ChatGPT scores and human scores was much more modest at the article level than at the journal level (Supplemental Figure 1). Coefficients of determination (R2) for linear regression ranged from 0.26 (general surgery) to 0.58 (obstetrics and gynecology). Likewise, global analysis of relevance scores assigned across all specialties revealed a clear but much weaker relation between human and ChatGPT-assigned relevance scores based on the standard error of the regression (Supplemental Figure 2). The standard error of the regression at the article level was 22.3, which is 2.5-fold greater than the journal-level analysis. We conclude that ChatGPT has only modest ability to classify the relevance of individual articles to specific domains of medicine.
Sensitivity and Quality Analyses
We visually inspected the distribution of scores for quality, accuracy, and bias produced by individual human reviewers, human reviewers in aggregate, and ChatGPT (Supplemental Figure 3). Score distributions were broadly similar, suggesting that harmonization instructions (Supplemental Appendix 1) given to reviewers were largely effective at standardizing scores of various human reviewers, without obvious variability due to individual reviewer leniency.
DISCUSSION
We evaluated whether the GPT-3.5 model, implemented as ChatGPT, could summarize medical research abstracts and determine the relevance of these articles to various medical specialties. Our analyses reveal that ChatGPT can produce high-quality, high-accuracy, and low-bias summaries of abstracts despite being given a word limit. We conclude that because ChatGPT summaries were 70% shorter than abstracts and usually of high quality, high accuracy, and low bias, they are likely to be useful as a screening tool to help busy clinicians and scientists more rapidly evaluate whether further review of an article is likely to be worthwhile. In Supplemental Appendix 3, we describe software—pyJournal-Watch—that might enable this kind of application.27-31 Life-critical medical decisions should for obvious reasons remain based on full, critical, and thoughtful evaluation of the full text of articles in context with available evidence from meta-analyses and professional guidelines. Our data also show that ChatGPT was much less able to classify the relevance of specific articles to various medical specialties. We had hoped to build a digital agent with the goal of consistently surveilling the medical literature, identifying relevant articles of interest to a given specialty, and forwarding them to a user. Chat-GPT’s inability to reliably classify the relevance of specific articles limits our ability to construct such an agent. We hope that in future iterations of LLMs, these tools will become more capable of relevance classification.
We are not aware of prior studies that have systematically evaluated GPT-3.5’s ability to summarize medical abstracts with a focus on quality, accuracy, and bias. However, our data are concordant with prior evidence suggesting reasonable performance for summarization in other domains (eg, news).32,33 Contrary to our expectations that hallucinations would limit the utility of ChatGPT for abstract summarization, this occurred in only 2 of 140 abstracts and was mainly limited to small (but important) methodologic or result details. Serious inaccuracies were likewise uncommon, occurring only in a further 2 of 140 articles. We conclude that ChatGPT summaries have rare but important inaccuracies that preclude them from being considered a definitive source of truth. Clinicians are strongly cautioned against relying solely on ChatGPT-based summaries to understand study methods and study results, especially in high-risk situations. Likewise, we noted at least 1 example in which the summary introduced bias by omitting gender as a significant risk factor in a logistic regression model, whereas all other significant risk factors were reported. In addition, concerns have been reasonably raised regarding biases inherent in LLMs.22,23 Here, we investigated whether—if carefully prompted—ChatGPT could nevertheless be used to produce low-bias summaries despite this known theoretical limitation.
The present study has limitations. First, we considered only a limited number of journals, and all abstracts focused on clinical medicine. Summarization performance on biomedical research at earlier stages of translational research (eg, articles describing fundamental mechanisms of cellular biology or biochemistry) was not evaluated by our analysis. We also focused exclusively on primary research reports, systematic reviews, and meta-analyses. We did not evaluate the performance of ChatGPT on abstracts from many other article types that are important to the scientific process including nonsystematic reviews, perspectives, commentaries, and letters to the editor. Second, because most journals now use a structured abstract, we included a small number of unstructured abstracts in our data set. Although we found no difference in ChatGPT performance in summarizing structured vs unstructured abstracts, it could be that a sample including more unstructured abstracts might detect performance differences with smaller effect sizes. Third, although we included journals with a broad range of impact factors (4.4-158.5), our analyses focused mostly on high-impact journals or journals that are particularly well regarded in their own specialty. Abstracts written for high-quality journals might be easier (or harder) to summarize than articles published in lower-tier journals. Performance in lower-impact journals could be interrogated in future studies. Fourth, all articles were chosen using simple random sampling except that we belatedly realized articles from Nature Medicine were sampled based on the order in which they were exported from PubMed rather than fully randomized. Sensitivity analyses excluding Nature Medicine did not change our conclusions; therefore, we kept articles from Nature Medicine in our analyses.
Large language models will continue to improve in quality. As we were finishing our analysis of ChatGPT based on the GPT-3.5 model, OpenAI began a limited beta release of the next generation in the GPT models, GPT-4. We suspect that as these models improve, summarization performance will be preserved and continue to improve. In addition, because the ChatGPT model was trained on pre-2022 data, it is possible that its slightly out-of-date medical knowledge decreased its ability to produce summaries or to self-assess the accuracy of its own summaries. In Supplemental Appendix 3, we report software that allows clinicians and scientists to immediately begin using GPT-3.5 and GPT-4 to systematically and rapidly review the clinical literature augmented by the advances in LLMs evaluated in this article. As LLMs evolve, future analyses should determine whether further iterations of the GPT language models have better performance in classifying the relevance of individual articles to various domains of medicine. In addition, in our analyses, we did not provide the LLMs with any article metadata such as the journal title or author list. Future analyses might investigate how performance varies when these metadata are provided. We encourage robust discussion within the family medicine research and clinical community on the responsible use of AI LLMs in family medicine research and primary care practice.
Footnotes
Conflicts of interest: authors report none.
Funding support: This work was not directly funded but used the REDCap data management platform at the University of Kansas Medical Center, which was supported by a Clinical and Translational Science Awards grant from the National Center for Advancing Translational Sciences (NCATS) awarded to the University of Kansas for Frontiers: University of Kansas Clinical and Translational Science Institute (#UL1TR002366). This work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or NCATS. This agency had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
- Received for publication April 21, 2023.
- Revision received October 13, 2023.
- Accepted for publication November 17, 2023.
- © 2024 Annals of Family Medicine, Inc.