Abstract
BACKGROUND Urinary tract infection (UTI) symptoms are common in primary care, but antibiotics are appropriate only when an infection is present. Urine culture is the reference standard test for infection, but results take >1 day. A machine learning predictor of urine cultures showed high accuracy for an emergency department (ED) population but required urine microscopy features that are not routinely available in primary care (the NeedMicro classifier).
METHODS We redesigned a classifier (NoMicro) that does not depend on urine microscopy and retrospectively validated it internally (ED data set) and externally (on a newly curated primary care [PC] data set) using a multicenter approach including 80,387 (ED) and 472 (PC) adults. We constructed machine learning models using extreme gradient boosting (XGBoost), artificial neural networks, and random forests (RFs). The primary outcome was pathogenic urine culture growing ≥100,000 colony forming units. Predictor variables included age; gender; dipstick urinalysis nitrites, leukocytes, clarity, glucose, protein, and blood; dysuria; abdominal pain; and history of UTI.
RESULTS Removal of microscopy features did not severely compromise performance under internal validation: NoMicro/XGBoost receiver operating characteristic area under the curve (ROC-AUC) 0.86 (95% CI, 0.86-0.87) vs NeedMicro 0.88 (95% CI, 0.87-0.88). Excellent performance in external (PC) validation was also observed: NoMicro/RF ROC-AUC 0.85 (95% CI, 0.81-0.89). Retrospective simulation suggested that NoMicro/RF can be used to safely withhold antibiotics for low-risk patients, thereby avoiding antibiotic overuse.
CONCLUSIONS The NoMicro classifier appears appropriate for PC. Prospective trials to adjudicate the balance of benefits and harms of using the NoMicro classifier are appropriate.
INTRODUCTION
Urinary tract infections (UTIs) are the most common type of infections managed in the outpatient setting, accounting for 1% to 3% of all consultations, 15% of all community prescriptions for antibiotics, and $1.6 billion in annual health care costs.1,2 Both men and women can be affected. Women have a 50% to 60% lifetime risk of UTI. Women >65 years of age are affected twice as often as the general female population.1 Surveys indicate that primary care (PC) clinicians are concerned about antibiotic resistance, but many view this as a public health issue in general rather than as a factor in prescribing decisions for individual patients.3
Urinary tract infection is usually diagnosed by combining history and physical examination with urine dipstick testing (including nitrite and leukocyte esterase).4,5 Microscopic evaluation of the urine to identify the presence of, for example, bacteria, leukocytes, and squamous epithelial cells is sometimes performed but is not always immediately available in the outpatient setting.
Urine culture is the reference standard for UTI diagnosis; however, urine cultures often take ≥24 hours for results whereas antibiotic treatment decisions are often made in minutes—during an office visit—at the point of care. Accurate prediction of urine cultures could enable prompt treatment of patients with UTI while avoiding antibiotic overuse for those without UTI.
Several approaches to more accurate diagnosis and treatment of UTI have been developed.6-11 Little et al7 developed a dipstick rule—based on the presence of nitrite or both leukocytes and blood—with a sensitivity of 77%, a specificity of 70%, and a negative predictive value (NPV) of 65%. Likewise, McIsaac et al8 developed a 3-variable decision aid (dysuria, leukocytes, nitrites) with a sensitivity of 80.3% and a specificity of 53.7%.
More recently, machine learning algorithms have been devised to predict the outcome of urine cultures.6,11 Heckerling et al6 used artificial neural networks (ANNs)12 to produce 5-variable predictors of urine culture results for a small data set (212 women). In a more recent study—using a much larger data set of >80,000 emergency department (ED) encounters—Taylor et al11 were able to predict the pathogenicity of a urine culture with high discriminative performance (reported receiver operating characteristic area under the curve [ROC-AUC] of 0.904 for the full [but impractical] 212-variable model and 0.877 for the reduced [but practical] 10-variable model). Their predictor leveraged a new machine learning approach based on extreme gradient boosting (XGBoost).11,13 The Taylor approach modeled the presence of a urine culture as a function of the following 10 variables: 2 demographic features (age, gender/sex), 3 urine dipstick features (nitrites, leukocyte esterase, blood), 2 history features (presence of dysuria, history of UTI), and 3 urine microscopy features (bacteria, epithelial cells, leukocytes).11
Unfortunately, in many ambulatory PC settings (eg, family medicine offices or urgent care facilities), urine microscopy is not immediately available, and treatment decisions are often made without this information. Urine microscopy provides valuable information for the evaluation of the pathogenicity of a urine culture. The presence of white blood cells and bacteria argues in favor of infection. Detection of squamous epithelial cells is a quality-control marker that suggests contamination with commensal urogenital flora. A risk therefore exists that removal of microscopic features from the prediction model might severely compromise performance.
We investigated whether the Taylor11 model could be adapted to remove the dependence on urine microscopy without compromising predictive accuracy and whether a model built on ED encounters could be generalized to PC patients at a different medical center. To that end, we developed a new model (NoMicro) that does not depend on urine microscopy variables. We trained and internally validated this new model on the original ED data set and then externally validated it on a new data set of 472 outpatient encounters in a family medicine office at a different institution.
METHODS
Data Sources
We used the following 2 data sources: a sample of >80,000 patients seen in an ED and previously described by Taylor et al11 (the ED data set), and a sample of 472 patients seen at the outpatient family medicine department at the University of Kansas Medical Center (the PC data set). Data extraction and quality assurance for the PC data set is detailed in Supplemental Appendix 1. The ED data set was further divided into training (80%, n = 64,310) and internal validation (20%, n = 16,077) data sets. The PC data set was used exclusively for external validation (ie, not for training). Characteristics of these data sets are summarized in Table 1.
Data Source Demographic Characteristics
Model Specification and Training
We trained urine culture predictive models using R v.3.6.1 (The R Foundation) using software and methods described by Taylor et al.11 The microscopy-required (NeedMicro) model was the Taylor et al11 model, specified as follows:
NeedMicro model, pathogenic culture = Age + Gender + (Nitrite * Leukocytes) + Blood + Dysuria + History of UTIs + (Microscopic bacteria * Microscopic epithelial cells) + Microscopic white blood cells
The microscopy-independent (NoMicro) model, using only data likely to be available during a PC office visit, was specified as follows:
NoMicro model, pathogenic culture = Age + Gender + Nitrite + Leukocytes + Blood + Clarity + Glucose + Protein + Dysuria + Abdominal pain + History of UTIs
To compensate for the loss of microscopy information, we added 4 features to the NoMicro model that might be (positively or negatively) associated with UTI, compared with relevant differential diagnoses, including 3 dipstick urinalysis features (clarity, glucose, protein) and 1 history feature (abdominal pain). These features can all be readily measured during a PC office visit.
We trained the NoMicro models using XGBoost,13 random forests (RFs),14,15 and ANNs.12 The NeedMicro model has been shown to perform best using XGBoost, and we trained this model using XGBoost as described.11
Model Validation
We internally validated trained models using the emergency department 20% holdout validation set and externally validated on the primary care data set. First, we determined the overall discriminative performance (ROC-AUC) and scaled Brier score. Clinical use of the classifiers depends not on their overall performance but on their performance at specific cutoffs for prediction of pathogenic and nonpathogenic; above the cutoff, cultures are predicted to be pathogenic, and below the cutoff, cultures are predicted to be non pathogenic. By varying the cutoff, greater sensitivity can be achieved in exchange for less specificity (and vice versa). We therefore characterized the sensitivity, specificity, positive predictive value (PPV), NPV, positive likelihood ratio, negative likelihood ratio, and diagnostic odds ratio at the optimal cutoff (ie, the cutoff maximizing the Youden index [sensitivity + specificity − 1]) and at a 15% false-negative rate (85% sensitivity [Sen85]). The significance of the 15% false-negative rate cutoff is that it allows for understanding of how the model will perform when used to reliably infer the absence of a pathogenic culture, as might be useful in supporting a decision to defer empiric antibiotic use (thereby decreasing antibiotic overuse). Finally, we determined model calibration; for example, a culture with a pathogenicity prediction of 30% should turn out to actually be pathogenic approximately 30% of the time. Further details of model validation can be found in Supplemental Appendix 1 Methods.16
Retrospective Evaluation of Potential to Decrease Antibiotic Overuse
We retrospectively evaluated the clinical effect of applying the following decision rules to the NoMicro models on the PC data set:
Rule 1. For patients with a culture predicted to be nonpathogenic under the Sen85 cutoff, we simulated the effect of withholding antibiotics.
Rule 2. For patients for whom the model predicts a pathogenic culture (at the Sen85 cutoff), we left the provision of antibiotics to physician discretion.
Our approach was to identify situations in which the models suggest antibiotics might be safely deferred to decrease antibiotic overuse. To mimic the population of a prospective clinical trial as closely as possible, we included all nonpregnant adults and excluded any individuals with high-risk features (eg, those suggestive of sepsis or pyelonephritis; see Supplemental Appendix 1).
Data and Source Code Availability
The deidentified PC data set and the statistical analysis code are available at https://github.com/djparente/uti-ml.
Human Subjects Protection
The University of Kansas Medical Center Institutional Review Board approved this project.
RESULTS
Comparison Between Emergency Department and Primary Care Data Sets
Demographic features of the ED and PC data sets are shown in Table 1. The ED data set comprised 80,387 individuals, whereas the PC data set comprised 472 individuals. Cultures were slightly more likely to be nonpathogenic in the ED data set (77.3%) compared with the PC data set (72.9%). Relative to PC patients, ED patients were more commonly older (32.9% aged >65 years vs 21.2% for PC patients), male (31.0% vs 13.6%), and of Hispanic/Latine/Spanish ethnicity (21.6% vs 12.3%). Racial distributions were broadly similar, although with a greater proportion of non-White patients in the PC data set (55.2%) compared with the ED data set (44.5%). The distribution of demographic and model predictor variables, stratified by urine culture pathogenicity, are reported in Supplemental Table 1.
Internal Validation of the Redesigned Classifier to Eliminate Dependence on Microscopy Data
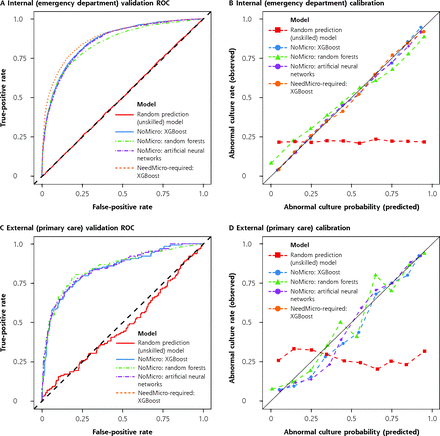
We compared the redesigned (NoMicro) classifier with the original (NeedMicro) classifier using the ED data set (internal validation). First, we evaluated overall performance (Table 2). The NoMicro classifier using XGBoost, RFs, and ANNs were evaluated against the best-performing NeedMicro classifier (which was trained using XGBoost11). For concision, we refer to, for example, the NoMicro classifier trained using the XGBoost algorithm as NoMicro/XGBoost. The 3 NoMicro classifiers performed similarly to each other and to the Need-Micro classifier (Table 2, Table 3, and Figure 1a). The best NoMicro classifier was achieved using XGBoost (as was the case for the NeedMicro classifier previously11). The ROC-AUC for the NoMicro/XGBoost classifier was 0.86 (95% CI, 0.86-0.87), and the NeedMicro classifier achieved an ROC-AUC of 0.88 (95% CI, 0.87-0.88; precisely in concordance with Taylor et al’s11 result).
Discriminative Performance (ROC-AUC), Calibration, and Brier Scores for the NoMicro and NeedMicro Predictive Models Under Internal (Emergency Department) and External (Primary Care) Validation
Cutoff-Varying Performance Metrics: Sensitivity, Specificity, Positive Predictive Value, Negative Predictive Value, Likelihood Ratios, and Diagnostic Odds Ratio
Discriminative performance and calibration of models under internal and external validation.
ROC = receiver operating characteristic; XGBoost = extreme gradient boosting.
Note: ROC (panels A and C) and calibration curves (panels B and D) for internal (emergency department, panels A-B) and external (primary care, panels C-D) validations. For internal validation, the NoMicro and NeedMicro models were evaluated. For external validation, only the NoMicro models were evaluated because microscopy is not routinely available in the primary care setting. Better models have ROC curves deflected away from the midline and toward the upper left corner. Well-calibrated models should lie along the diagonal line. The performance of unskilled classifiers (which return random results) were also simulated and are shown for comparison.
Next, we evaluated performance measures at 2 prediction cutoffs (Table 3). At the optimal cutoff, the best NoMicro classifier (NoMicro/XGBoost) achieved superior sensitivity (80.0%; 95% CI, 78.7%-81.3%) compared with the NeedMicro classifier (76.1%; 95% CI, 74.6%-77.5%), at the cost of specificity (NoMicro/XGBoost: 76.3%; 95% CI, 75.6%-77.1% vs NeedMicro: 83.7%; 95% CI, 83.0%-84.3%). At the Sen85 cutoff, the NoMicro and NeedMicro classifiers achieved similar specificity (NoMicro/XGBoost: 70.5%; 95% CI, 69.7%-71.3% vs NeedMicro: 73.1%; 95% CI, 72.4%-73.9%). At this threshold, both models also had excellent NPV (NoMicro/XGBoost: 94.3%; 95% CI, 93.9%-94.7% vs NeedMicro: 94.5%; 95% CI, 94.1%-94.9%).
We further evaluated the calibration of the models. All models were well calibrated on the ED validation data set (Table 2, Supplemental Table 2, and Figure 1b). Linear fits of the observed vs predicted pathogenicity rate within risk deciles (Supplemental Table 2) explained most of the variability in the decile plots (R2 ≥ 0.995 for all fits). All models—except NoMicro/RF—also produced decile-plot fits with slopes and intercepts approximately equal to 1 and 0, respectively, as expected. The pattern of residuals around the 45° perfect-calibration line in the decile plots also showed no systematic deviation, except in the case of NoMicro/RF, which underestimated the pathogenicity of predicted-to-be-pathogenic cultures and overestimated the pathogenicity of predicted-to-be-benign cultures.
External Validation on the Primary Care Data Set
We next determined whether the NoMicro classifiers would perform adequately in our clinical setting of interest (PC rather than ED) at a different institution (external validation). The NoMicro classifiers all performed excellently on this data set (Table 2, Table 3, and Figure 1c). The best NoMicro classifier, NoMicro/RF, achieved an overall ROC-AUC of 0.85 (95% CI, 0.81-0.89). At the optimal threshold, NoMicro/RF had a sensitivity of 78.9% (95% CI, 71.9%-85.2%) and a specificity of 81.4% (95% CI, 77.6%-85.5%). At the Sen85 threshold, NoMicro/RF had a specificity of 66.0% (95% CI, 60.8%-70.9%). Importantly, the NPV at the Sen85 threshold was 92.3% (95% CI, 89.1%-95.1%).
Regarding calibration, the pattern of fluctuations above and below the ideal calibration line (a line with slope 1 and intercept 0) showed no systemic deviation, indicating that none of the NoMicro models systematically overestimated or underestimated predictions. Indeed, all models—this time including NoMicro/RF—also produced decile-plot fits with slopes and intercepts approximately equal to 1 and 0, respectively, as expected, on the PC data set. However, within risk deciles, there was greater variability, with corresponding lower—but still high—R2 values (Table 2, R2 values from 0.94 to 0.98, compared with the ED data set, with R2 >0.99 for all models). Note that we could not evaluate the NeedMicro classifier on the PC data set because nearly all (~90%) PC encounters did not include microscopy data (indeed, this was the original rationale for the development of the NoMicro model).
Potential to Decrease Antibiotic Overuse
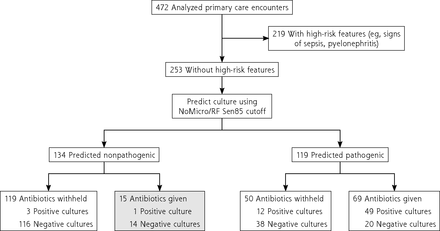
We evaluated whether the NoMicro model could be used to potentially decrease antibiotic overuse (Figure 2, Supplemental Figure 1, and Supplemental Figure 2). Of 472 included encounters, 219 had ≥1 high-risk feature, whereas 253 lacked high-risk features. Of the 253 individuals lacking these features, the NoMicro/RF model (Figure 2) predicted 119 cultures to be pathogenic and 134 to be nonpathogenic. In the predicted-pathogenic arm, the decision rule (Rule 2) recommends no change to antibiotic decision making. In the predicted-nonpathogenic arm, there were 15 instances in which the decision rule (Rule 1) would have recommended withholding antibiotics in situations in which physicians (without the benefit of the decision rule) prescribed antibiotics. Almost all decisions to prescribe antibiotics contrary to the decision rule’s recommendation to withhold them would have been incorrect; 14 of the 15 instances (93.3%) had negative urine cultures, whereas only 1 instance had a positive culture. Similar results were obtained using the other classifiers NoMicro/XGBoost (Supplemental Figure 1) and NoMicro/ANN (Supplemental Figure 2). For NoMicro/RF, the decision rules would have increased the incidence for which antibiotics were (correctly) withheld from patients with negative urine cultures from 81.9% to 89.4% (+7.4%) while only increasing the incidence for which antibiotics were (incorrectly) withheld from patients with positive urine cultures from 23.1% to 24.6% (+1.5%).
Evaluation of the potential of NoMicro to decrease antibiotic overuse (random forests).
RF = random forests; Sen85 = threshold obtained by requiring the greatest specificity such that sensitivity is >85% (ie, false-negative rate is <15%).
Note: Of 472 primary care encounters, 253 lacked high-risk features for progression to serious illness and were stratified using the NoMicro/Random Forests classifier at the Sen85 cutoff (false-negative rate 15%). These predictions were correlated with physician antibiotic prescribing behavior (made without the benefit of the NoMicro/RF model). The shaded box represents cases for which the NoMicro/RF model predicts the culture to be nonpathogenic but for which physicians nevertheless prescribed antibiotics; almost all cultures in this group were negative. Antibiotic overuse might be decreased by withholding antibiotics for this group.
DISCUSSION
We investigated whether a previously successful urine culture prediction model11 in an ED could be adapted to remove its dependence on microscopy data (the NoMicro model), thereby rendering it appropriate for environments that lack the ability to characterize urine microscopically at the point of care (eg, PC or urgent care). In internal validation (ED data set), a statistical difference between the ROC-AUC for the NoMicro/XGBoost and NeedMicro classifiers was found (DeLong test P < .001). However, although the large sample size allowed this statistical difference to be detected, this difference is unlikely to have major clinical implications; both the NoMicro and NeedMicro classifiers achieved high performance (ROC-AUC both >0.85). Likewise, cutoff-dependent performance measures (sensitivity, specificity, PPV, and NPV) were broadly comparable in the NoMicro and NeedMicro models. Taken together, the overall and cutoff-dependent performance under internal validation suggest that the NoMicro classifiers are viable alternatives to the NeedMicro classifier and are not severely impaired by the loss of urine microscopy features from the prediction model.
Similarly, performance statistics under external validation—in a different clinical setting (PC) at a different institution—were similar to those obtained during internal validation. This strongly suggests that removal of microscopic features does not substantially impair performance of the prediction model, the model is not significantly overfit to the peculiarities of the ED data set, and the use of the NoMicro classifiers in PC populations is reasonable and generalizable across at least some institutions and practice settings.
Specifically, our results establish the validity of the NoMicro model in a single-center ED (internal validation) and a different, but still single-center, PC environment (external validation) at an urban academic medical center. That the NoMicro model—which was trained on the ED data set—works well in a completely different clinical setting (PC) and physical location suggests that the model is generalizable. Our results nevertheless do not definitively establish this. It remains formally possible that the model might not be valid in settings with more profound differences (eg, an affluent, suburban, community urgent care). Future studies might investigate this.
At both the optimal and Sen85 cutoffs, the NPV of the NoMicro/RF model was excellent (>90%). However, the PPV was much lower (61.2%; 95% CI, 56.0%-67.3% at the optimal cutoff and 48.2%; 95% CI, 44.1%-52.6% at the Sen85 cutoff). Our model might therefore be useful for UTI in a manner that is analogous to how a d-dimer test might be used to rule out pulmonary embolism17; useful to withhold antibiotics (reasonably exclude infection) when negative but not useful to infer an infection when positive. (That is, antibiotics should not be started solely based on a pathogenic prediction from the NoMicro model). Our results therefore suggest that use of the proposed decision rules could decrease antibiotic overuse without substantially withholding antibiotics in the setting of an infection.
The NoMicro model is more complicated than scoring system–type decision rules that simply add up the number of risk factors present and compare them to a prespecified cutoff. Custom software—preferably web-based, for broad availability—will need to be written and validated to allow clinicians to act on NoMicro predictions. Importantly, such software should not describe predicted-pathogenic results as high risk or potentially pathogenic, which might unintentionally cause physicians to prescribe antibiotics when they otherwise might have chosen to defer them. Development and validation of such a tool is a major future direction of this work. Among the goals of such work would be to understand under what conditions the NoMicro predictions (the NoMicro model is essentially a black-box model) are viewed as acceptable to PC physicians.
The present study has limitations. First, the PC data set was relatively small (n = 472) and described a single center. The total number of individuals prescribed antibiotics, for whom the proposed decision rules suggested antibiotics be withheld, was correspondingly small (15 individuals). Second, the ED and PC data sets had a pathogenic urine culture prevalence of ~25%. Measured ROC-AUC, sensitivity, and specificity do not change with population prevalence, but NPV does. In a practice setting with a greater prevalence of pathogenic urine cultures, the NoMicro NPV will be lower. Third, our data are necessarily limited to cases in which a urine culture was ordered. Urine cultures are likely to be ordered in situations in which a patient is very sick, in which case the principle clinical need is to obtain speciation data and antibiotic sensitivities. However, urine cultures might also be ordered in cases in which a patient seems to be clinically stable, to defer treatment pending the culture, with the expectation that the culture will be negative. Focusing analysis on patients for whom a urine culture was ordered therefore likely introduces a bias into our analysis, but we are not sure how strongly (or in what direction) this bias influences our results.
Our present results suggest that future prospective evaluation of the proposed decision rules as a tool to decrease antibiotic overuse is justified and is unlikely to cause harm to nonpregnant adults without high-risk features.
Footnotes
Conflicts of interest: authors report none.
Funding support: This work was not directly funded but used the REDCap data management platform at the University of Kansas Medical Center, which was supported by a Clinical and Translational Science Awards grant from the National Center for Advancing Translational Sciences (NCATS) awarded to the University of Kansas for Frontiers: University of Kansas Clinical and Translational Science Institute (#UL1TR002366). This work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or NCATS. This agency had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; or decision to submit the manuscript for publication.
- Received for publication March 18, 2022.
- Revision received August 23, 2022.
- Accepted for publication August 31, 2022.
- © 2023 Annals of Family Medicine, Inc.